 |
MRT811S- METHODS IN RECOMBINANT DNA TECHNOLOGY - 2ND Opp - JULY 2022 |
 |
1 Page 1 |
▲back to top |

gp E
NAMIBIA UNIVERSITY
OF SCIENCE AND TECHNOLOGY
FACULTY OF HEALTH, APPLIED SCIENCES AND NATURAL RESOURCES
DEPARTMENT OF NATURAL AND APPLIED SCIENCES
QUALIFICATION: BACHELOR OF SCIENCE HONOURS
QUALIFICATION CODE: O8BOSH
LEVEL: 8
COURSE CODE: MRT811S
COURSE NAME: METHODS IN RECOMBINANT DNA
TECHNOLOGY
SESSION: JULY 2022
DURATION: 3 HOURS
PAPER: THEORY
MARKS: 100
EXAMINER
SECOND OPPORTUNITY QUESTION PAPER
DR LAMECH MWAPAGHA
MODERATOR | DR RONNIE BOCK
INSTRUCTIONS
Answer ALL the questions.
Write clearly and neatly.
Number the answers clearly.
All written work MUST be done in BLUE or BLACK ink.
PERMISSIBLE MATERIALS
None
THIS QUESTION PAPER CONSISTS OF FOUR (4) PAGES
(Including this front page)
Page 1 of4
 |
2 Page 2 |
▲back to top |

Question 1
[11]
a) Discuss the mode of action of small interfering RNAs (siRNAs)
(3)
b) Messenger RNA caps are made in three steps, each performed by a different enzyme. Name
the THREE (3) enzymes.
(3)
c) A Bachelor of Science research student would like to knock-out a gene of interest in a
gene expression study using the CRISPR/Cas9 technique. Discuss in detail the mechanism
behind this technique.
(5)
Question 2
[12]
a) Briefly describe the Blue-white screening as a rapid and efficient technique for the identification of
recombinant bacteria.
(3)
b) Describe FOUR (4) limitations of blue-white screening
(4)
c) ERA1, a gene identified in Arabidopsis, which is involved in Abscisic acid (ABA) signaling. ABA, a
plant stress hormone, induces the closure of leaf stomata thereby reducing water loss through
transpiratioann,d decreasing the rate of photosynthesis. Thus, plants lacking ERA1 activity have
increased tolerance to drought. Discuss in details how one would go about cloning the ERA1 gene
in a bacterial plasmid.
(5)
Question 3
[15]
a) Give SIX (6) Similarities between the Ti and Ri Plasmids
(6)
b) Mention THREE (3) Limitations to the particle bombardment method, compared to
Agrobacterium-mediated transformation
(3)
c) What are the steps followed in plant transformation using particle bombardment?
(6)
Question 4
[14]
a) What are the functions of the following genes as utilised in genetic engineering?
(3)
l. Reporter gene;
Il. Marker gene;
Ill. Selectable marker;
Page 2 of4
 |
3 Page 3 |
▲back to top |
b) The Cre-Lox system relies on two components to function, a Cre recombinase and its recognition
site loxP. Based on the orientation of the loxP sites, describe the THREE (3) outcomes that can
result from the recombination.
(6)
c) Describe the Cre Recombinase mechanism
(5)
Question 5
[18]
a) Explain the following medical applications of transgenic animals.
(9)
|. | Xenotransplanters;
Il. | Tools in the study of immunoglobulin genes;
lll. | Chemical Safety Testing;
b) There are several approaches in which scientific researchers may use in correcting faulty genes.
Briefly describe FOUR (4) of these approaches
(4)
c) Gene therapy is a medical approach that treats or prevents disease by correcting the underlying
genetic problem. What are some of the risks envisioned in Gene Therapy?
(5)
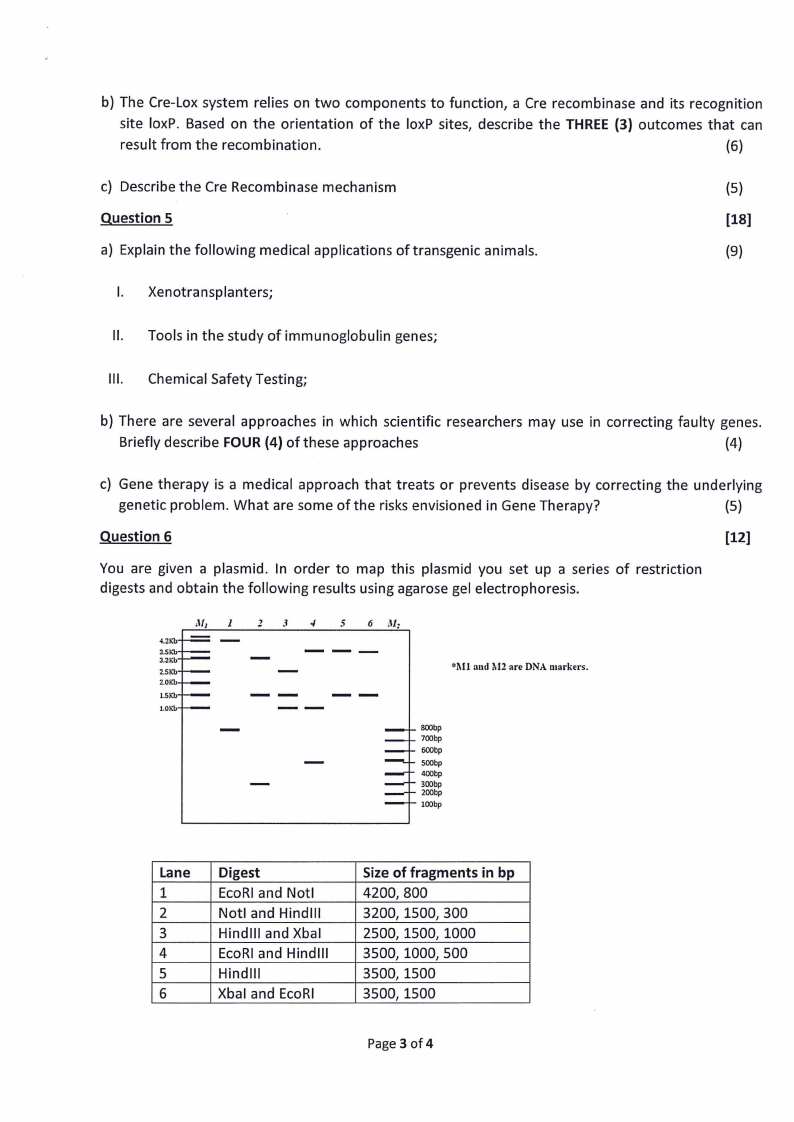
Question 6
(12]
You are given a plasmid. In order to map this plasmid you set up a series of restriction
digests and obtain the following results using agarose gel electrophoresis.
M,
I
2
3
4
3
4.2Kb
33..25KKbb7]]
22..50KKbb--]4
LSkb4
1.0Kb>
ones
ae
ens
6 = M;
*N{1 and M2 are DNA markers.
emmm-|—
wee}
moame——
~=m——
emme-
=——=—T-
ae—
———
800bp
700bp
G600bp
SOObp
400bp
300bp
+200bp
100bp
Lane
Digest
1
EcoRI and Notl
Z
Notl and Hindill
3
Hindlll and Xbal
4
EcoRI and Hindlll
5
Hindlll
6
Xbal and EcoRI
Size of fragments in bp
4200, 800
3200, 1500, 300
2500, 1500, 1000
3500, 1000, 500
3500, 1500
3500, 1500
Page 3 of4
 |
4 Page 4 |
▲back to top |

a) What is the approximate size of the plasmid?
(2)
b) Add the Notl, Hindlll, Xbal restriction sites onto the plasmid map shown below. On your map give
the distances between each of the restriction sites.
(5)
EcoRI
c) Mention FIVE (5) factors that make a Vector ideal for cloning.
(5)
Question 7
[18]
a) The genetic code is the set of rules defining how the four-letter code of DNA is translated into
amino acids, which are the building blocks of proteins. Discuss THREE (3) characteristics of the
genetic code
(6)
b) Briefly describe the following mutations;
(8)
|. |Nonsense Mutation;
ll. Frame shift Mutation;
Il. = Silent Mutation;
IV. Misense Mutation;
c) State FOUR (4) benefits of using mice in transgenic studies?
(4)
THE END
Page 4 of 4





