 |
MOD621S - MOLECULAR DIAGNOSTICS - 2ND OPP - JAN 2023 |
 |
1 Page 1 |
▲back to top |

n Am I BI A u n IVER s ITY
OF SCIEn CE Ano TECH n OLOGY
FACULTYOF HEALTH,APPLIEDSCIENCESAND NATURAL RESOURCES
DEPARTMENT OF HEALTH SCIENCES
QUALIFICATION: BACHELOR OF MEDICAL LABORATORY SCIENCES
QUALIFICATION CODE: 08BMLS
LEVEL: 6
COURSE CODE: MOD621S
COURSE NAME: MOLECULAR DIAGNOSTICS
SESSION:
JANUARY 2023
·'
PAPER:
THEORY
DURATION:
3 HOURS
MARKS:
100
SUPPLEMENTARY/SECONDOPPORTUNITY EXAMINATION PAPER
EXAMINER(S) Ms. V. Tjijenda
MODERATOR: Dr A Shiningavamwe
INSTRUCTIONS
1. Answer ALL the questions.
2. Write clearly and neatly.
3. Number the answers clearly.
PERMISSIBLEMATERIALS
Scientific Calculator
THIS MEMORANDUM CONSISTS OF 5 PAGES (Including this front page)
 |
2 Page 2 |
▲back to top |
SECTION A (10)
QUESTION 1
[10]
Evaluate the statements in each numbered section and indicate whether the
statement is true or false. Write "true 11 or "false 11 next to the corresponding
number and correct each false statement.
1.1 SYBRGreen can bind to primer dimer including the desired target sequence
during amplification.
1.2 T4 Polynucleatide Kinase is used for the addition of a hydroxyl group to an
end having a free phosphate group.
1.3 The melting temperature of the CGGAGATTCTAGACCTCCTGis 66 °c.
1.4 During electrophoresis, high-molecular-weight DNA migrates slower
than low-molecular-weight DNA through an agarose gel?
1.5 In preparing 250ml of a 0.8 % agarose gel, 0.8 g agarose is dissolved in
250ml TAE buffer.
1.6 Increasing the Mg 2+ concentration in a PCRmixture increases the activity
of Taq Polymerase and its specificity.
SECTION B (40)
QUESTION 2
[17]
2.1 When performing any conventional PCRit is important to include negative
and positive controls. Explain what negative and positive controls are, and
why is it necessary to include them.
(6)
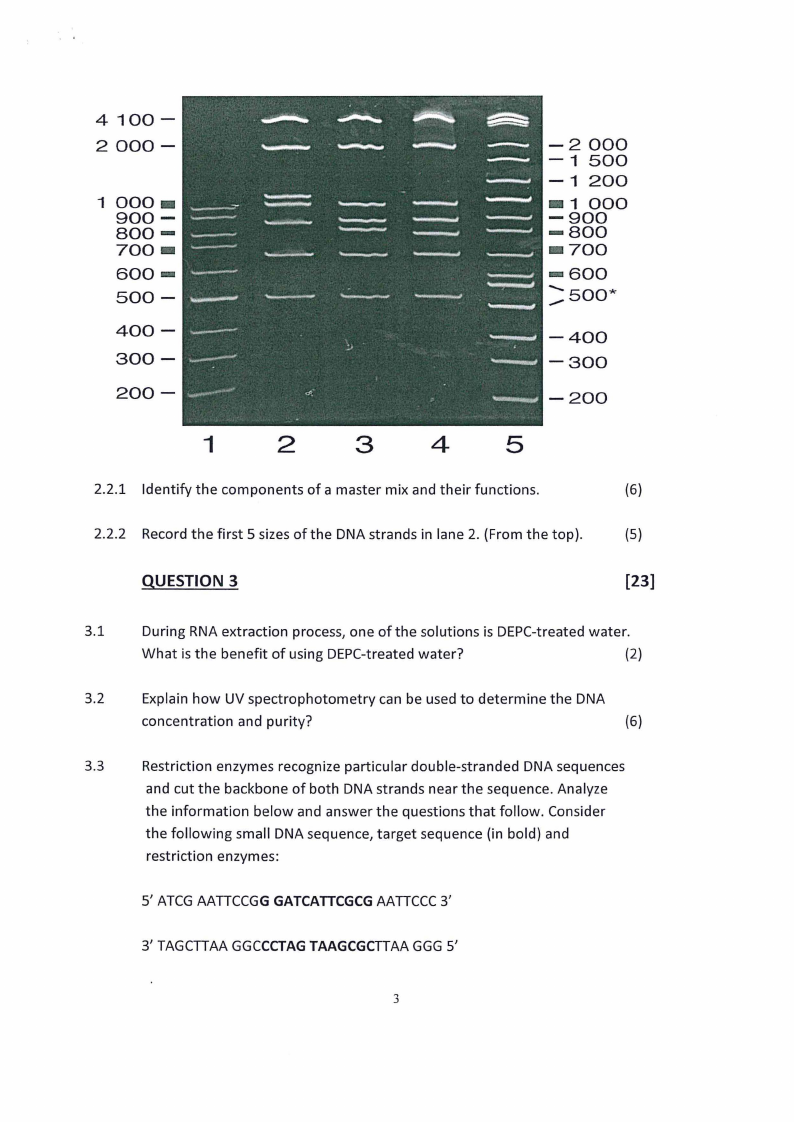
2.2 Study the image below and answer the questions that follow:
2
 |
3 Page 3 |
▲back to top |
4 100-
2 000-
1 ooo.
900-
800-
700•
600-
500-
400-
300-
200-
-2 000
-1 500
-1 200
•1 000
--9s0o0 o
•700
--·-600
.____5,00*
-400
-300
-200
1
2
3
4
5
2.2.1 Identify the components of a master mix and their functions.
(6)
2.2.2 Record the first 5 sizes of the DNA strands in lane 2. {From the top).
{5)
QUESTION 3
[23]
3.1
During RNA extraction process, one of the solutions is DEPC-treated water.
What is the benefit of using DEPC-treated water?
(2)
3.2
Explain how UV spectrophotometry can be used to determine the DNA
concentration and purity?
(6)
3.3
Restriction enzymes recognize particular double-stranded DNA sequences
and cut the backbone of both DNA strands near the sequence. Analyze
the information below and answer the questions that follow. Consider
the following small DNA sequence, target sequence {in bold) and
restriction enzymes:
5' ATCGAATTCCGGGATCATTCGCGAATTCCC3'
3' TAGCTTAA GGCCCTAGTAAGCGCTTAAGGG 5'
3
 |
4 Page 4 |
▲back to top |

Enzyme
EcoRI
Barn HI
Mbol
Target sequence (cut at*) 5'-----+3'
G*AATTC
G*GATCC
*GATC
3.3.1 Define palindromic sequence.
(2)
3.3.2 For each of the restriction enzymes listed, give the number of times that
the enzymes will cut the DNA fragment above. Also give the number of
resulting DNA fragments after individual treatment with each enzyme. (6)
3.3.3 Calculate the Tm of the sense strand.
(4)
3.3.4 Which enzyme will you use to clone the target sequence?
(1)
3.3.5 Provide two reasons why you prefer the enzyme mention in 3.3.4 for
plasmid cloning?
(2)
SECTIONC (SO)
QUESTION 4
[20]
4.1 Describe four technical problems that can occur during the different steps
in the Northern Blot Hybridisation method resulting in failure of a signal to
appear on the final blot.
(5)
4.2 Describe how the FISHtechnique is used to diagnose Philadelphia
Chromosome.
(7)
4.3 Discuss 'Nested Primer PCR'in detail. Under what circumstance would this
type of PCRmethod be adopted and explain its benefit.
(8}
4
 |
5 Page 5 |
▲back to top |

QUESTION 5
[30]
5.1 Discuss the steps in Western Blotting technique.
{10)
5.2 Summarize the main steps involved in setting up and running gel
Electrophoresis.
{10)
5.3.1 Define sequencing.
{2)
5.3.2 Differentiate between Sanger sequencing and Maxam Gilbert.
{4)
5.3.3 Below is the gel profile obtained from sequencing a gene using sanger
sequencing. Provide the sequence of interest.
{4)
t--
-
-
END OF EXAMINATION!
5





